Anne L. Martel
Breast Cancer Detection from Multi-View Screening Mammograms with Visual Prompt Tuning
Apr 28, 2025Abstract:Accurate detection of breast cancer from high-resolution mammograms is crucial for early diagnosis and effective treatment planning. Previous studies have shown the potential of using single-view mammograms for breast cancer detection. However, incorporating multi-view data can provide more comprehensive insights. Multi-view classification, especially in medical imaging, presents unique challenges, particularly when dealing with large-scale, high-resolution data. In this work, we propose a novel Multi-view Visual Prompt Tuning Network (MVPT-NET) for analyzing multiple screening mammograms. We first pretrain a robust single-view classification model on high-resolution mammograms and then innovatively adapt multi-view feature learning into a task-specific prompt tuning process. This technique selectively tunes a minimal set of trainable parameters (7\%) while retaining the robustness of the pre-trained single-view model, enabling efficient integration of multi-view data without the need for aggressive downsampling. Our approach offers an efficient alternative to traditional feature fusion methods, providing a more robust, scalable, and efficient solution for high-resolution mammogram analysis. Experimental results on a large multi-institution dataset demonstrate that our method outperforms conventional approaches while maintaining detection efficiency, achieving an AUROC of 0.852 for distinguishing between Benign, DCIS, and Invasive classes. This work highlights the potential of MVPT-NET for medical imaging tasks and provides a scalable solution for integrating multi-view data in breast cancer detection.
Enhancing breast cancer detection on screening mammogram using self-supervised learning and a hybrid deep model of Swin Transformer and Convolutional Neural Network
Apr 28, 2025Abstract:Purpose: The scarcity of high-quality curated labeled medical training data remains one of the major limitations in applying artificial intelligence (AI) systems to breast cancer diagnosis. Deep models for mammogram analysis and mass (or micro-calcification) detection require training with a large volume of labeled images, which are often expensive and time-consuming to collect. To reduce this challenge, we proposed a novel method that leverages self-supervised learning (SSL) and a deep hybrid model, named \textbf{HybMNet}, which combines local self-attention and fine-grained feature extraction to enhance breast cancer detection on screening mammograms. Approach: Our method employs a two-stage learning process: (1) SSL Pretraining: We utilize EsViT, a SSL technique, to pretrain a Swin Transformer (Swin-T) using a limited set of mammograms. The pretrained Swin-T then serves as the backbone for the downstream task. (2) Downstream Training: The proposed HybMNet combines the Swin-T backbone with a CNN-based network and a novel fusion strategy. The Swin-T employs local self-attention to identify informative patch regions from the high-resolution mammogram, while the CNN-based network extracts fine-grained local features from the selected patches. A fusion module then integrates global and local information from both networks to generate robust predictions. The HybMNet is trained end-to-end, with the loss function combining the outputs of the Swin-T and CNN modules to optimize feature extraction and classification performance. Results: The proposed method was evaluated for its ability to detect breast cancer by distinguishing between benign (normal) and malignant mammograms. Leveraging SSL pretraining and the HybMNet model, it achieved AUC of 0.864 (95% CI: 0.852, 0.875) on the CMMD dataset and 0.889 (95% CI: 0.875, 0.903) on the INbreast dataset, highlighting its effectiveness.
A generalizable 3D framework and model for self-supervised learning in medical imaging
Jan 20, 2025



Abstract:Current self-supervised learning methods for 3D medical imaging rely on simple pretext formulations and organ- or modality-specific datasets, limiting their generalizability and scalability. We present 3DINO, a cutting-edge SSL method adapted to 3D datasets, and use it to pretrain 3DINO-ViT: a general-purpose medical imaging model, on an exceptionally large, multimodal, and multi-organ dataset of ~100,000 3D medical imaging scans from over 10 organs. We validate 3DINO-ViT using extensive experiments on numerous medical imaging segmentation and classification tasks. Our results demonstrate that 3DINO-ViT generalizes across modalities and organs, including out-of-distribution tasks and datasets, outperforming state-of-the-art methods on the majority of evaluation metrics and labeled dataset sizes. Our 3DINO framework and 3DINO-ViT will be made available to enable research on 3D foundation models or further finetuning for a wide range of medical imaging applications.
VertDetect: Fully End-to-End 3D Vertebral Instance Segmentation Model
Nov 16, 2023



Abstract:Vertebral detection and segmentation are critical steps for treatment planning in spine surgery and radiation therapy. Accurate identification and segmentation are complicated in imaging that does not include the full spine, in cases with variations in anatomy (T13 and/or L6 vertebrae), and in the presence of fracture or hardware. This paper proposes VertDetect, a fully automated end-to-end 3D vertebral instance segmentation Convolutional Neural Network (CNN) model to predict vertebral level labels and segmentations for all vertebrae present in a CT scan. The utilization of a shared CNN backbone provides the detection and segmentation branches of the network with feature maps containing both spinal and vertebral level information. A Graph Convolutional Network (GCN) layer is used to improve vertebral labelling by using the known structure of the spine. This model achieved a Dice Similarity Coefficient (DSC) of 0.883 (95% CI, 0.843-0.906) and 0.882 (95% CI, 0.835-0.909) in the VerSe 2019 and 0.868 (95\% CI, 0.834-0.890) and 0.869 (95\% CI, 0.832-0.891) in the VerSe 2020 public and hidden test sets, respectively. This model achieved state-of-the-art performance for an end-to-end architecture, whose design facilitates the extraction of features that can be subsequently used for downstream tasks.
Understanding metric-related pitfalls in image analysis validation
Feb 09, 2023Abstract:Validation metrics are key for the reliable tracking of scientific progress and for bridging the current chasm between artificial intelligence (AI) research and its translation into practice. However, increasing evidence shows that particularly in image analysis, metrics are often chosen inadequately in relation to the underlying research problem. This could be attributed to a lack of accessibility of metric-related knowledge: While taking into account the individual strengths, weaknesses, and limitations of validation metrics is a critical prerequisite to making educated choices, the relevant knowledge is currently scattered and poorly accessible to individual researchers. Based on a multi-stage Delphi process conducted by a multidisciplinary expert consortium as well as extensive community feedback, the present work provides the first reliable and comprehensive common point of access to information on pitfalls related to validation metrics in image analysis. Focusing on biomedical image analysis but with the potential of transfer to other fields, the addressed pitfalls generalize across application domains and are categorized according to a newly created, domain-agnostic taxonomy. To facilitate comprehension, illustrations and specific examples accompany each pitfall. As a structured body of information accessible to researchers of all levels of expertise, this work enhances global comprehension of a key topic in image analysis validation.
Metrics reloaded: Pitfalls and recommendations for image analysis validation
Jun 03, 2022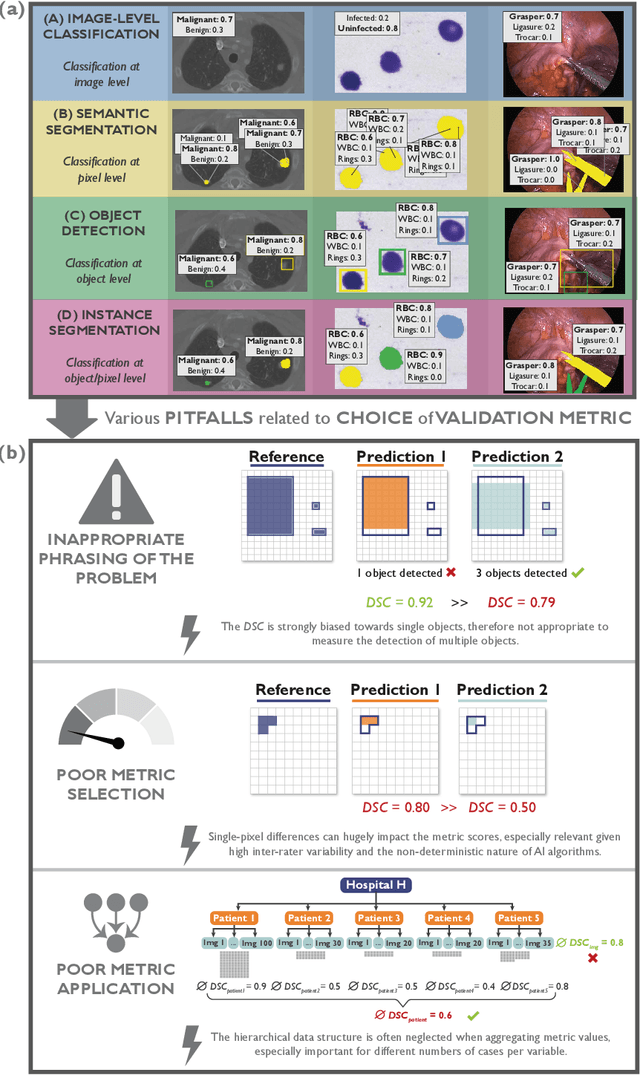
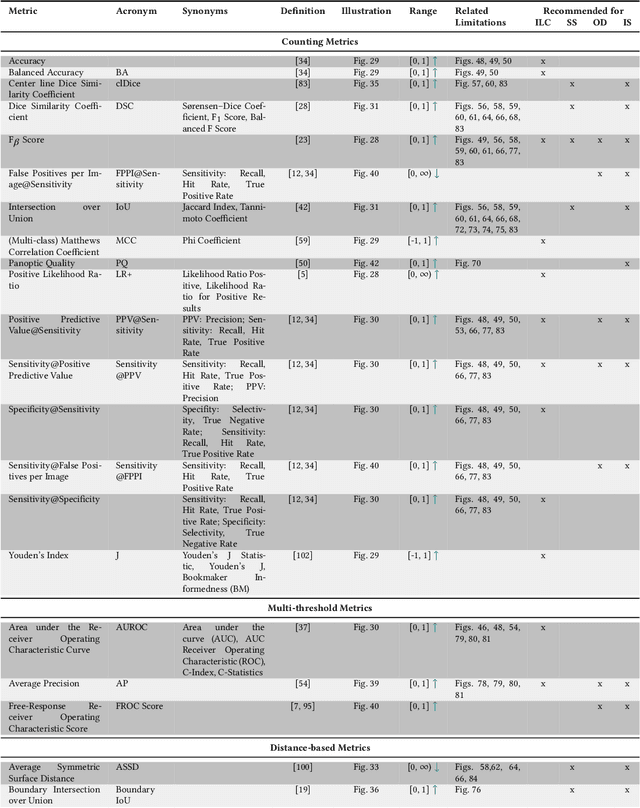
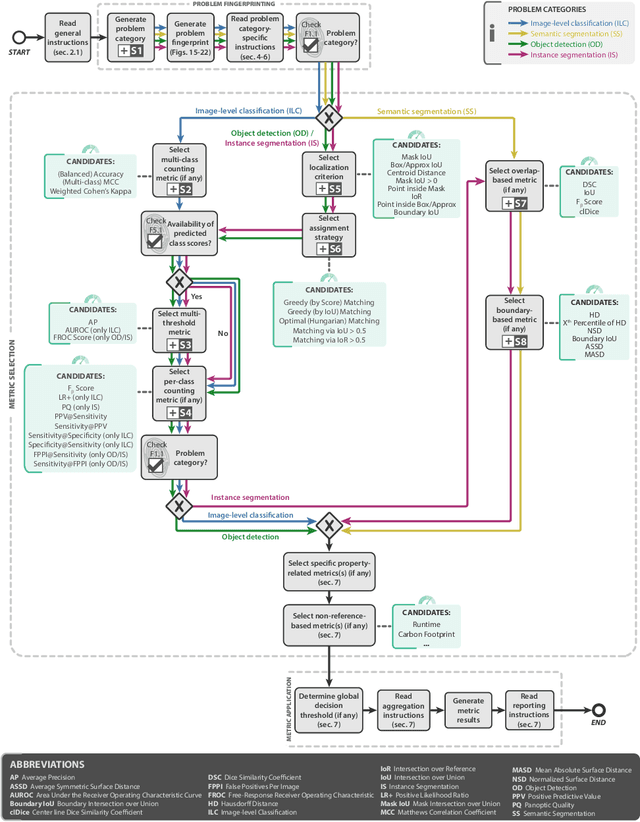
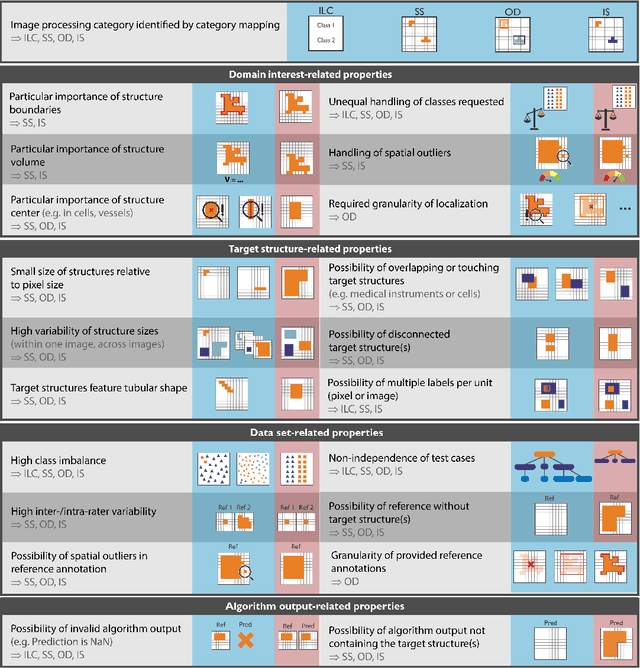
Abstract:The field of automatic biomedical image analysis crucially depends on robust and meaningful performance metrics for algorithm validation. Current metric usage, however, is often ill-informed and does not reflect the underlying domain interest. Here, we present a comprehensive framework that guides researchers towards choosing performance metrics in a problem-aware manner. Specifically, we focus on biomedical image analysis problems that can be interpreted as a classification task at image, object or pixel level. The framework first compiles domain interest-, target structure-, data set- and algorithm output-related properties of a given problem into a problem fingerprint, while also mapping it to the appropriate problem category, namely image-level classification, semantic segmentation, instance segmentation, or object detection. It then guides users through the process of selecting and applying a set of appropriate validation metrics while making them aware of potential pitfalls related to individual choices. In this paper, we describe the current status of the Metrics Reloaded recommendation framework, with the goal of obtaining constructive feedback from the image analysis community. The current version has been developed within an international consortium of more than 60 image analysis experts and will be made openly available as a user-friendly toolkit after community-driven optimization.
ROOD-MRI: Benchmarking the robustness of deep learning segmentation models to out-of-distribution and corrupted data in MRI
Mar 11, 2022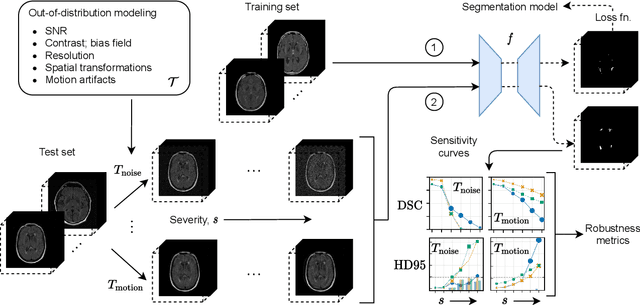
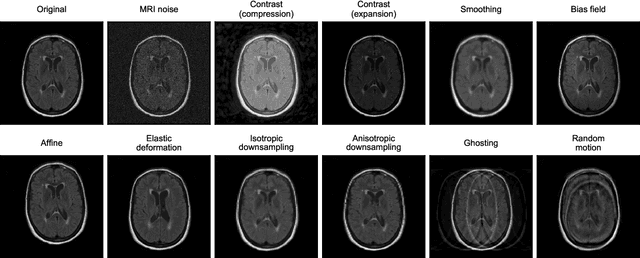
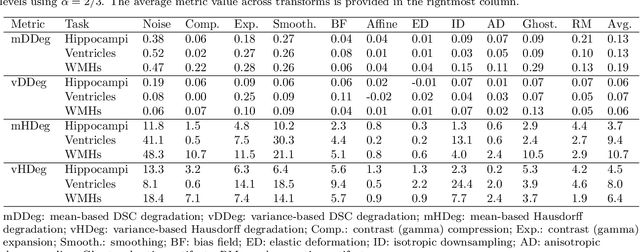
Abstract:Deep artificial neural networks (DNNs) have moved to the forefront of medical image analysis due to their success in classification, segmentation, and detection challenges. A principal challenge in large-scale deployment of DNNs in neuroimage analysis is the potential for shifts in signal-to-noise ratio, contrast, resolution, and presence of artifacts from site to site due to variances in scanners and acquisition protocols. DNNs are famously susceptible to these distribution shifts in computer vision. Currently, there are no benchmarking platforms or frameworks to assess the robustness of new and existing models to specific distribution shifts in MRI, and accessible multi-site benchmarking datasets are still scarce or task-specific. To address these limitations, we propose ROOD-MRI: a platform for benchmarking the Robustness of DNNs to Out-Of-Distribution (OOD) data, corruptions, and artifacts in MRI. The platform provides modules for generating benchmarking datasets using transforms that model distribution shifts in MRI, implementations of newly derived benchmarking metrics for image segmentation, and examples for using the methodology with new models and tasks. We apply our methodology to hippocampus, ventricle, and white matter hyperintensity segmentation in several large studies, providing the hippocampus dataset as a publicly available benchmark. By evaluating modern DNNs on these datasets, we demonstrate that they are highly susceptible to distribution shifts and corruptions in MRI. We show that while data augmentation strategies can substantially improve robustness to OOD data for anatomical segmentation tasks, modern DNNs using augmentation still lack robustness in more challenging lesion-based segmentation tasks. We finally benchmark U-Nets and transformer-based models, finding consistent differences in robustness to particular classes of transforms across architectures.
Metastatic Cancer Outcome Prediction with Injective Multiple Instance Pooling
Mar 09, 2022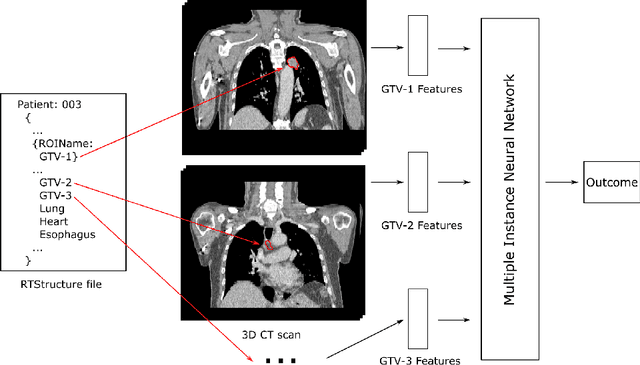
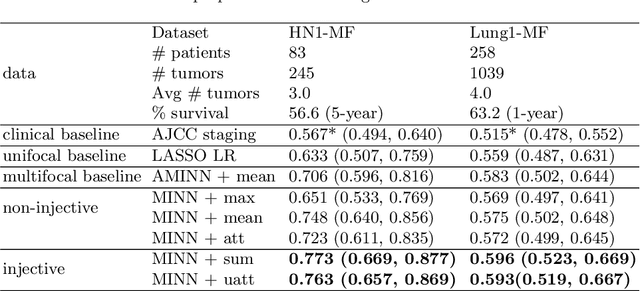
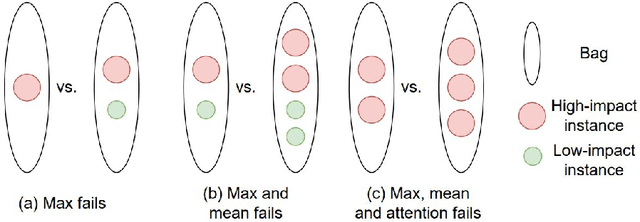
Abstract:Cancer stage is a large determinant of patient prognosis and management in many cancer types, and is often assessed using medical imaging modalities, such as CT and MRI. These medical images contain rich information that can be explored to stratify patients within each stage group to further improve prognostic algorithms. Although the majority of cancer deaths result from metastatic and multifocal disease, building imaging biomarkers for patients with multiple tumors has been a challenging task due to the lack of annotated datasets and standard study framework. In this paper, we process two public datasets to set up a benchmark cohort of 341 patient in total for studying outcome prediction of multifocal metastatic cancer. We identify the lack of expressiveness in common multiple instance classification networks and propose two injective multiple instance pooling functions that are better suited to outcome prediction. Our results show that multiple instance learning with injective pooling functions can achieve state-of-the-art performance in the non-small-cell lung cancer CT and head and neck CT outcome prediction benchmarking tasks. We will release the processed multifocal datasets, our code and the intermediate files i.e. extracted radiomic features to support further transparent and reproducible research.
BI-RADS BERT & Using Section Tokenization to Understand Radiology Reports
Oct 14, 2021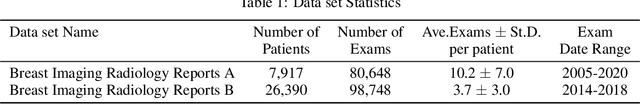

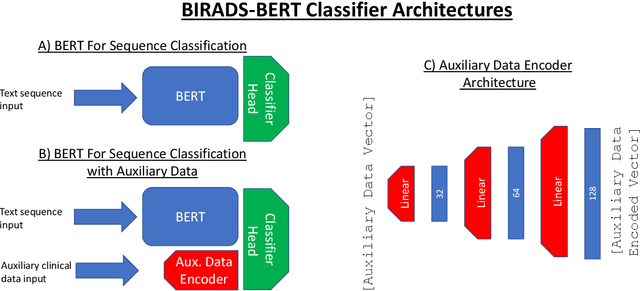
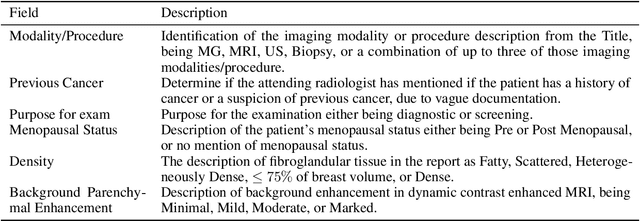
Abstract:Radiology reports are the main form of communication between radiologists and other clinicians, and contain important information for patient care. However in order to use this information for research it is necessary to convert the raw text into structured data suitable for analysis. Domain specific contextual word embeddings have been shown to achieve impressive accuracy at such natural language processing tasks in medicine. In this work we pre-trained a contextual embedding BERT model using breast radiology reports and developed a classifier that incorporated the embedding with auxiliary global textual features in order to perform a section tokenization task. This model achieved a 98% accuracy at segregating free text reports into sections of information outlined in the Breast Imaging Reporting and Data System (BI-RADS) lexicon, a significant improvement over the Classic BERT model without auxiliary information. We then evaluated whether using section tokenization improved the downstream extraction of the following fields: modality/procedure, previous cancer, menopausal status, purpose of exam, breast density and background parenchymal enhancement. Using the BERT model pre-trained on breast radiology reports combined with section tokenization resulted in an overall accuracy of 95.9% in field extraction. This is a 17% improvement compared to an overall accuracy of 78.9% for field extraction for models without section tokenization and with Classic BERT embeddings. Our work shows the strength of using BERT in radiology report analysis and the advantages of section tokenization in identifying key features of patient factors recorded in breast radiology reports.
Resource and data efficient self supervised learning
Sep 03, 2021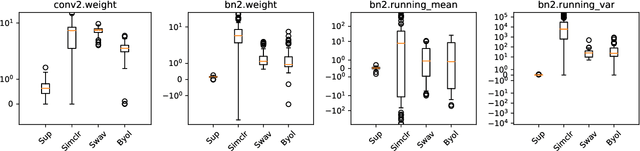

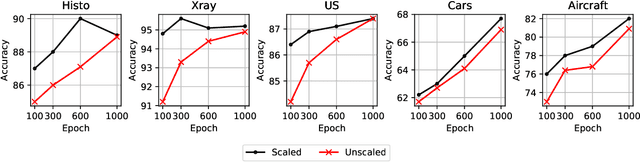
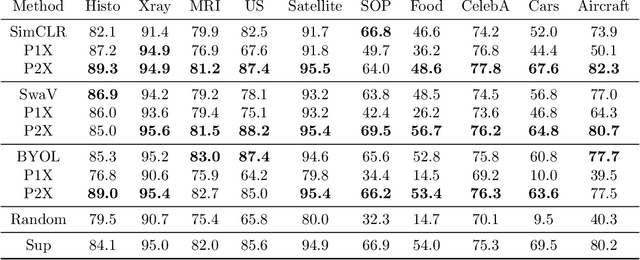
Abstract:We investigate the utility of pretraining by contrastive self supervised learning on both natural-scene and medical imaging datasets when the unlabeled dataset size is small, or when the diversity within the unlabeled set does not lead to better representations. We use a two step approach which is analogous to supervised training with ImageNet initialization, where we pretrain networks that are already pretrained on ImageNet dataset to improve downstream task performance on the domain of interest. To improve the speed of convergence and the overall performance, we propose weight scaling and filter selection methods prior to second step of pretraining. We demonstrate the utility of this approach on three popular contrastive techniques, namely SimCLR, SWaV and BYOL. Benefits of double pretraining include better performance, faster convergence, ability to train with smaller batch sizes and smaller image dimensions with negligible differences in performance. We hope our work helps democratize self-supervision by enabling researchers to fine-tune models without requiring large clusters or long training times.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge